Start of topic | Skip to actions
First Provenance Challenge
Aims
The provenance challenge aims to establish an understanding of the capabilities of available provenance-related systems and, in particular, the following details.- The representations that systems use to document details of processes that have occurred
- The capabilities of each system in answering provenance-related queries
- What each system considers to be within scope of the topic of provenance (regardless of whether the system can yet achieve all problems in that scope)
- Representations of the workflow in their system
- Representations of provenance for the example workflow
- Representations of the result of the core (and other) queries
- Contributions to a matrix of queries vs systems, indicating for each that: (1) the query can be answered by the system, (2) the system cannot answer the query now but considers it relevant, (3) the query is not relevant to the project.
- Additional queries (beyond the core queries) that illustrate the scope of their system
- Extensions to the example workflow to best illustrate the unique aspects of their system
- Any categorisation of queries that the project considers to have practical value
Example Workflow
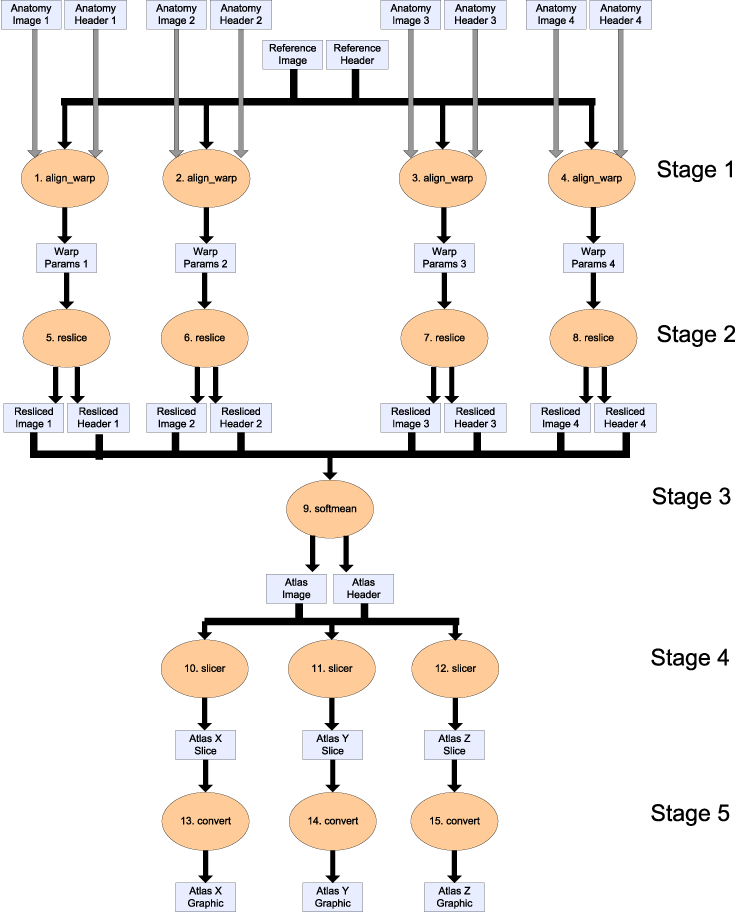
We propose an example workflow for creating population-based "brain atlases" from the fMRI Data Center's archive of high resolution anatomical data. The workflow is shown below (click for a pdf version of the image). It is comprised of procedures, shown as orange ovals, and data items flowing between them, shown as rectangles. It can be seen as five stages, where each stage is depicted as a horizontal row of the same procedure in the figure. Note that the term stage is introduced only to help description of the workflow, and we do not dictate how it is apparent in a concrete implementation. The procedures employ the AIR (automated image registration) suite to create an averaged brain from a collection of high resolution anatomical data, and the FSL suite to create 2D images across each sliced dimension of the brain. In addition to the data items shown in the figure, there are other inputs to procedures (constant string options), defined below. The inputs to a workflow are a set of new brain images (Anatomy Image 1 to 4) and a single reference brain image (Reference Image). All input images are 3D scans of a brain of varying resolutions, so that different features are evident. For each image, there is the actual image and the metadata information for that image (Anatomy Header 1 to 4). The image data was published with article Frontal-Hippocampal Double Dissociation Between Normal Aging and Alzheimer's Disease by Head, D, Synder, AZ, Girton, LE, Morris, JC, Buckner, RL in the fMRI Data Center Accession Number: 2-2004-1168X. The stages of the workflow are as follows.- For each new brain image, align_warp compares the reference image to determine how the new image should be warped, i.e. the position and shape of the image adjusted, to match the reference brain. The output of each procedure in the stage is a _warp parameter set_ defining the spatially transformation to be performed (Warp Params 1 to 4).
- For each warp parameter set, the actual transformation of the image is done by reslice, which creates a new version of the original new brain image with the configuration defined in the warp parameter set. The output is a resliced image.
- All the resliced images are averaged into one single image using softmean.
- For each dimension (x, y and z), the averaged image is sliced to give a 2D atlas along a plane in that dimension, taken through the centre of the 3D image. The output is an atlas data set, using slicer. This tool can be downloaded as part of the FSL suite, available at http://www.fmrib.ox.ac.uk/fsl/.
- For each atlas data set, it is converted into a graphical atlas image using (the ImageMagick utility) convert.
Core Provenance Queries
An initial set of provenance-related queries is given below.- Find the process that led to Atlas X Graphic / everything that caused Atlas X Graphic to be as it is. This should tell us the new brain images from which the averaged atlas was generated, the warping performed etc.
- Find the process that led to Atlas X Graphic, excluding everything prior to the averaging of images with softmean.
- Find the Stage 3, 4 and 5 details of the process that led to Atlas X Graphic.
- Find all invocations of procedure align_warp using a twelfth order nonlinear 1365 parameter model (see model menu describing possible values of parameter "-m 12" of align_warp) that ran on a Monday.
- Find all Atlas Graphic images outputted from workflows where at least one of the input Anatomy Headers had an entry global maximum=4095. The contents of a header file can be extracted as text using the scanheader AIR utility.
- Find all output averaged images of softmean (average) procedures, where the warped images taken as input were align_warped using a twelfth order nonlinear 1365 parameter model, i.e. "where softmean was preceded in the workflow, directly or indirectly, by an align_warp procedure with argument -m 12."
- A user has run the workflow twice, in the second instance replacing each procedures (convert) in the final stage with two procedures: pgmtoppm, then pnmtojpeg. Find the differences between the two workflow runs. The exact level of detail in the difference that is detected by a system is up to each participant.
- A user has annotated some anatomy images with a key-value pair center=UChicago. Find the outputs of align_warp where the inputs are annotated with center=UChicago.
- A user has annotated some atlas graphics with key-value pair where the key is studyModality. Find all the graphical atlas sets that have metadata annotation studyModality with values speech, visual or audio, and return all other annotations to these files.
Participant Instructions
We here give the specific steps that we expect each participating team to perform in completing the challenge.- The partipant should determine how they are going to execute the workflow (or a simulation of it) and how it will record data (provenance) about the execution.
- The team should add the provenance to their TWiki page, and to declare the way in which they executed the workflow, e.g. upload a workflow script.
- If the partipant has varied the workflow to make it more suitable for their system or to demonstrate an aspect important to their approach, then they should declare what this variation is.
- The team should then use their systems to answer the core provenance queries, and any others that they wish to perform to demonstrate key aspects of their system.
- The participant then uploads to the TWiki the queries performed, the way in which the queries were expressed/realised, and the answers they got.
- For core queries that were not performed, the partipant should say why they were not performed, i.e. whether the query is considered out of scope for the system or in scope but not currently possible to answer.
- For any data given above, each team should provide a link to an explanation of the representation used so that other participants can interpret it.
Sample Workflow Implementations
As it may be useful to some, we provide sample implementations of the workflow here. This should not preclude the use of any other technology. The implementations assume that the executables referenced above are all installed; they are provided by the two packages AIR (automated image registration) suite and ImageMagick. Minor caution - this is a DOS text file, and if run on Unix the extra carriage returns at the ends of lines make their way into the filenames and cause everything to break. Strip the CRs with tr before running...Timetable
- 2006-June: Challenge finalised, participants start!
- 2006-September-13: Deadline for challenge results to be uploaded
- 2006-September-13 and 2006-September-14: Face-to-face meeting at which results are discussed
- 2006-October-15: Comparisons performed, minutes of discussion, proposed next steps uploaded
to top
| I | Attachment  | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|
| | BrainAtlas.png | manage | 5.1 K | 16 May 2006 - 15:02 | SimonMiles | Brain Atlas workflow (original vdt display) |
| | BrainAtlas.pdf | manage | 118.8 K | 30 May 2006 - 16:40 | SimonMiles | Brain Atlas workflow (hi-res) |
| | workflow.sh | manage | 0.8 K | 31 May 2006 - 15:07 | SimonMiles | Shell script version of workflow |
| | BrainAtlas.gif | manage | 40.1 K | 06 Jun 2006 - 17:39 | LucMoreau |